Compare the flu pandemic of 1918 and COVID-19 with caution – the past is not a prediction

A pandemic from a century ago doesn’t necessarily chart the course of the pandemic happening now. National Photo Company Collection/Library of Congress Prints and Photographs Division/Library of Congress Prints and Photographs Division, CC BY
Courtesy of Mari Webel, University of Pittsburgh and Megan Culler Freeman, University of Pittsburgh
People have turned to historical experience with influenza pandemics to try to make sense of COVID-19, and for good reason.
Influenza and coronavirus share basic similarities in the way they’re transmitted via respiratory droplets and the surfaces they land on. Descriptions of H1N1 influenza patients in 1918-19 echo the respiratory failure of COVID-19 sufferers a century later. Lessons from efforts to mitigate the spread of flu in 1918-19 have justifiably guided this pandemic’s policies promoting nonpharmaceutical interventions, such as physical distancing and school closures.
Current discussions about scaling back social distancing measures and “opening up” the country frequently refer to “waves” of disease that characterized the dramatic mortality of H1N1 influenza in three major peaks in 1918-19. As COVID-19 rates begin to steady in some parts of the U.S., people today are nervously eyeing the “second wave” of influenza that came in autumn 1918, that pandemic’s deadliest period.
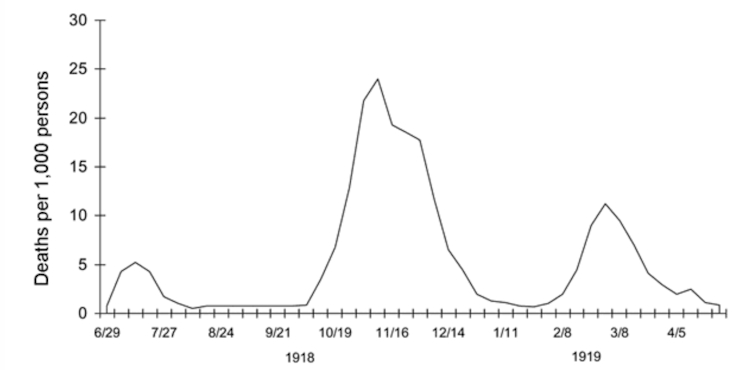
Waves evoke predictability, however, and COVID-19 has been hard to predict. Despite the valuable lessons drawn from past influenza outbreaks, how pandemic influenza struck in 1918 isn’t a template for what will happen with COVID-19 in the coming months.
As a historian and a virologist, we believe this comparison of two pandemics has contributed to public confusion about what to expect from “flattening the curve.” Key divergences in the sociopolitical contexts of 1918-19 and now, in addition to clear virologic differences between influenza and SARS-CoV-2, the virus that causes COVID-19, mean their courses are not perfectly matched.
Influenza pandemic a product of that time
Today’s citizens may consider the 2020 world to be dramatically more connected than in the past. But World War I and soldier mobilization created a situation well-suited to influenza dispersal. While the origin of the deadly strain of 1918 H1N1 remains obscure, evidence indicates that soldiers on the move drove circulation.
Young American men left their homes – rural farms, small towns, crowded cities – and traveled around the world. They gathered by the thousands in military training camps and on troop ships, and then at the front in Europe. Civilians globally continued to work in crucial areas of economic production that required movement through the same transit hubs soldiers used. The disease’s first wave occurred in spring and early summer 1918 amid these movements.
H1N1 flu stowed away with soldiers returning from World War I. Keystone View Co./Library of Congress Prints and Photographs Division, CC BY
In theaters of war in Europe, Africa and western Asia, soldiers mingled with their global compatriots. When they demobilized, they passed through major transit hubs back to their homes around the world, interacting with more people.
The extraordinarily deadly second wave of influenza in autumn 1918 diffused linearly along rail and sea routes, then radiated outward to wreak havoc on previously unexposed populations globally. In some areas, this period was followed by a less deadly third winter wave of disease in early 1919.
Medical historians conservatively estimate that influenza killed 50 million people globally, with 675,000 in the United States between 1918 and 1920. After that, this strain of flu receded, likely due to changes in the virus itself and the fact that most people had already been exposed and developed immunity or died.
Because the waves of pandemic flu did recede, it’s tempting to imagine today’s pandemic following a similar trajectory. However, fundamental differences between the biology of SARS-CoV-2 and influenza viruses make it hard to chart the future of COVID-19 based on what happened in the early 20th century.
SARS-CoV-2 and flu are biologically different
Both the new coronavirus and influenza have genetic material in the form of RNA. RNA viruses tend to accumulate a lot of mutations as they multiply – they typically don’t double-check copied genes to correct errors during replication. These mutations can occasionally lead to significant changes: The virus might change the species it infects or cell receptor it uses, or it could become more or less deadly, or spread more or less easily.
Uniquely, influenza’s genetic material is organized in segmented chunks. This idiosyncrasy means the virus can trade entire segments of RNA with other influenza viruses, enabling rapid evolution. Influenza also has a distinct seasonality, circulating much more during the winter months. As virus strains circulate, oscillating seasonally between the Northern and Southern Hemispheres’ wintertimes, they mutate rapidly. This capacity for quick adaptation is why you need to get a new flu vaccination annually to protect against new strains that have emerged in your area since last year.
SARS-CoV-2 makes many copies of itself once it successfully infects a human cell. dowell/Moment via Getty Images
Coronaviruses actually do proofread their copied RNA to fix inadvertent errors during replication, which decreases their relative mutation rate. From the originally sequenced SARS-CoV-2 in Wuhan, China in December 2019 to recently banked sequences from the U.S., there are fewer than 10 mutations in 30,000 potential locations in its genome, despite the virus having traveled around the world and through multiple generations of human hosts. Influenza makes 6.5 times more errors per replication cycle, independent of entire genome segment swaps.
The relative genetic stability of SARS-CoV-2 means that future peaks of disease are unlikely to be driven by natural changes in virulence due to mutation. Mutation is unlikely to contribute to predictable “waves” of COVID-19.
It’s also currently unknown if SARS-CoV-2 will be influenced by the seasons, like influenza. It has already successfully spread in many climates. It’s hard to attribute recent declines in the rate of new cases to warmer weather – they’re occurring in the wake of various strict nonpharmaceutical interventions.
All this means that oscillations in COVID-19 cases are unlikely to come with the predictability that discussions of influenza “waves” in 1918-19 might suggest. Rather, as SARS-CoV-2 continues to circulate in nonimmune populations globally, physical distancing and mask-wearing will keep its spread in check and, ideally, keep infection and death rates steady.
As states loosen nonpharmaceutical interventions, the U.S. will likely experience a long plateau of continued new infections at a steady rate, punctuated by periodic local flares. These outbreaks will not be driven by SARS-CoV-2 mutation or virulence, but by the further exposure of nonimmune people to the virus. Future spikes in COVID-19 cases and deaths will very likely be driven by what people do.
This scenario will continue until the U.S. population gains herd immunity, ideally accelerated by vaccination. Unfortunately, this process may be measured in years rather than months.
One virus’s pattern is not a prediction
People seek answers from the experiences of influenza in 1918-19 for a fundamental reason: It ended.
History shows the pandemic ebbed after a final, third wave in spring 1919 without the benefit of an influenza vaccine (available only in the mid-1940s) or a molecular or serologic test, or effective antiviral therapy, or even the support of mechanical ventilation.
The earlier pandemic does hold lessons for the current one, including the value of wearing masks to stop the virus’s spread. Topical Press Agency/Hulton Archive via Getty Images
Today we’re living through a novel pandemic. By and large, people are actively collaborating in unprecedented measures to disrupt transmission of SARS-CoV-2. Scanning the historical record is one way to draw our own lives into focus and perspective. Unfortunately, the end of influenza in summer 1919 does not portend the end of COVID-19 in the summer of 2020.
The pandemic’s scientific complexities are formidable challenges. They’re playing out in a global economy that has ground to a halt, with resultant increasing pressures to reopen communities, and a technologically advanced and interconnected society – all issues that our predecessors a century ago did not have to consider.
Jessica Pickett, Ph.D., a principal consultant with Tomorrow Global, LLC, contributed to this article.
[Get facts about coronavirus and the latest research. Sign up for The Conversation’s newsletter.]
Mari Webel, Assistant Professor of History, University of Pittsburgh and Megan Culler Freeman, Pediatric Infectious Diseases Fellow, University of Pittsburgh
![]()
This article is republished from The Conversation under a Creative Commons license. Read the original article.